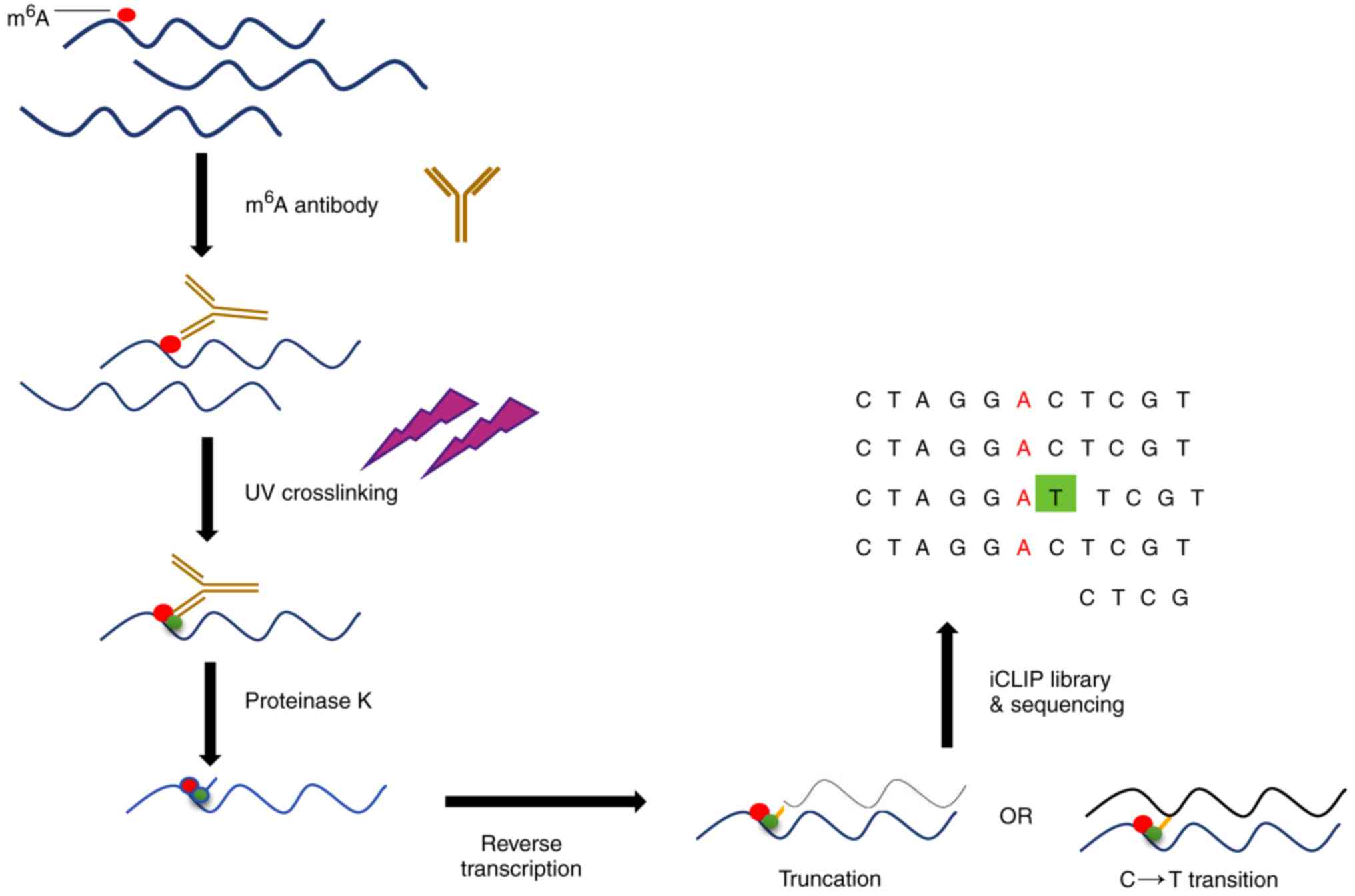
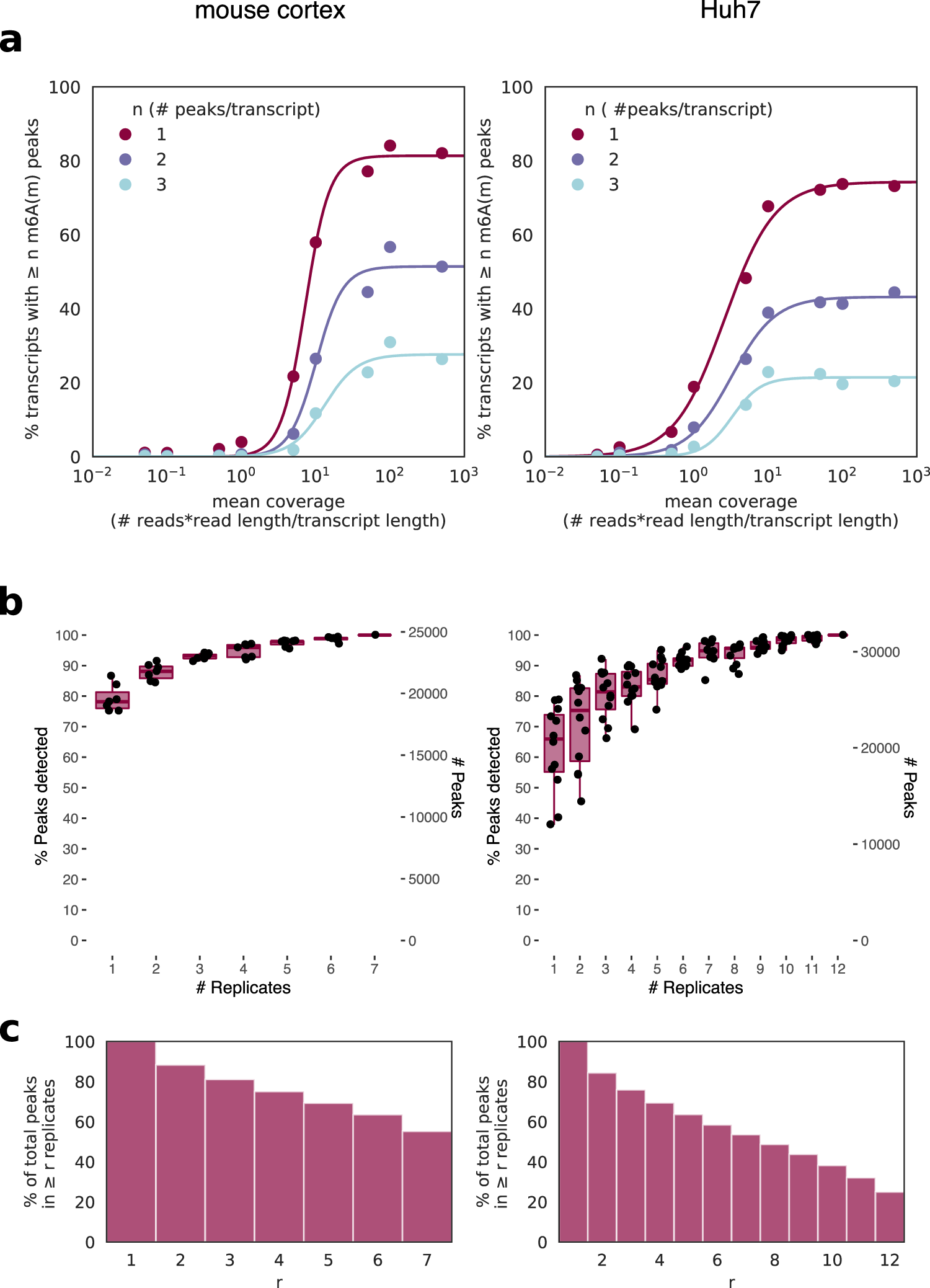
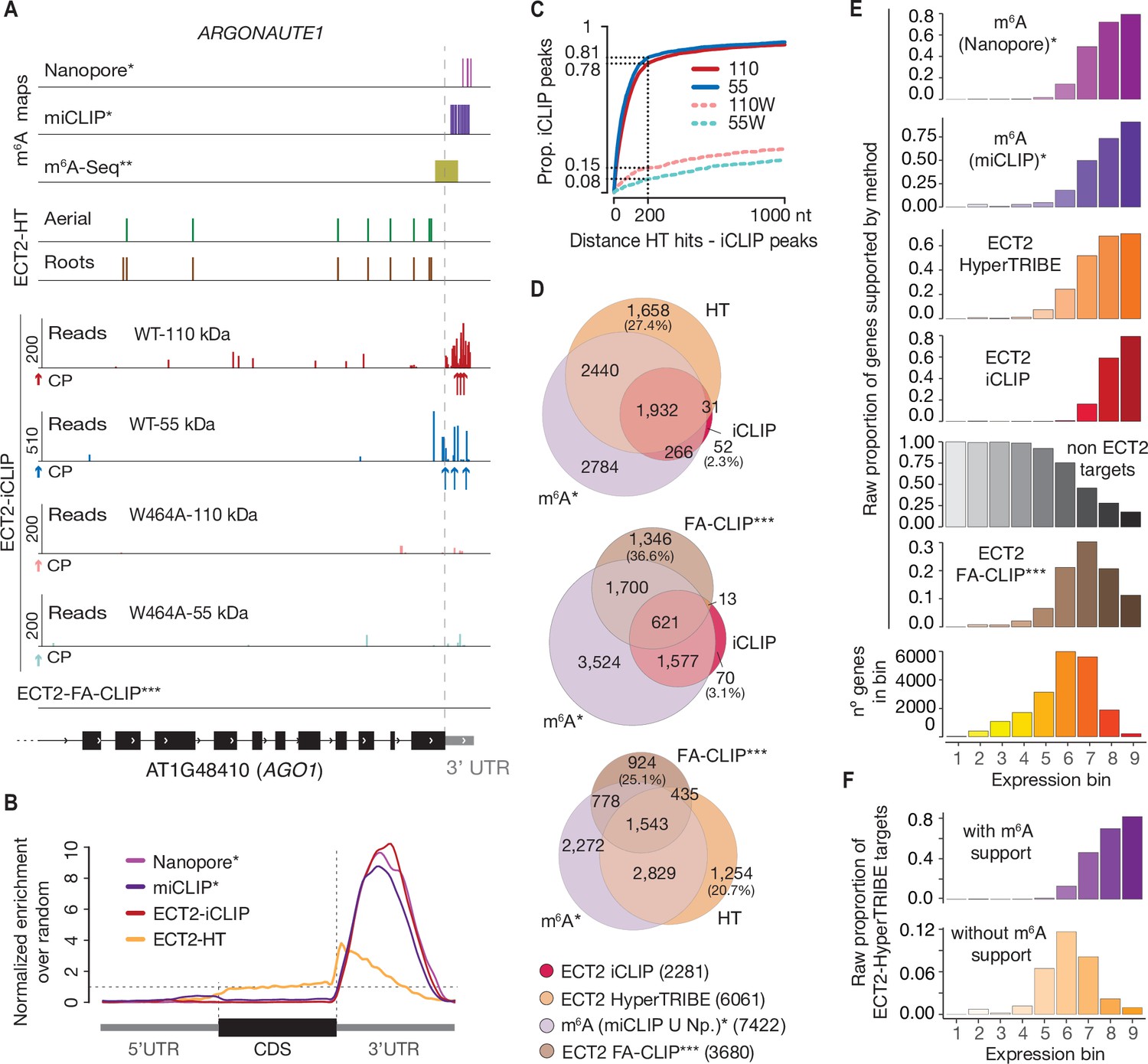
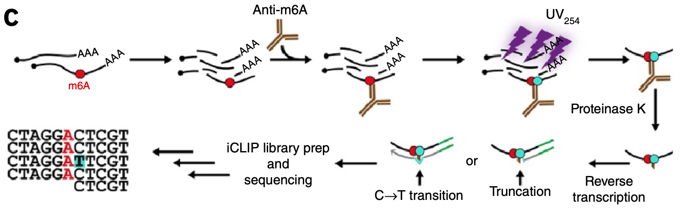
![PDF] High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. | Semantic Scholar PDF] High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/e1a071ac484a9f518fb5a1fe468ffeaa06250139/3-Figure2-1.png)
PDF] High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. | Semantic Scholar

METTL3 and WTAP bind m6A consensus sequence in mRNA and affect gene... | Download Scientific Diagram

Evolutionary conservation of the DRACH signatures of potential N6-methyladenosine (m6A) sites among influenza A viruses | Scientific Reports

m6A reader Pho92 is recruited co-transcriptionally and couples translation efficacy to mRNA decay to promote meiotic fitness in yeast | bioRxiv

Down-Regulation of m6A mRNA Methylation Is Involved in Dopaminergic Neuronal Death | ACS Chemical Neuroscience
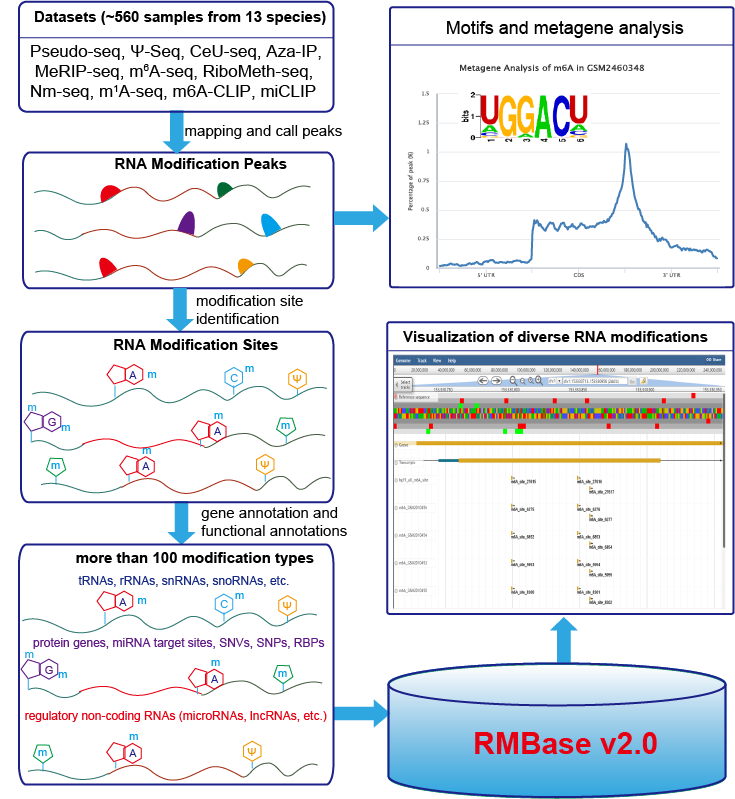
RNA Modification Database, RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data (Pseudo-seq, Ψ-seq, CeU-seq, Aza-IP, MeRIP-seq, m6A-seq, RiboMeth-seq, m1A-seq).

Relevance of N6-methyladenosine regulators for transcriptome: Implications for development and the cardiovascular system - Journal of Molecular and Cellular Cardiology

N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. – 武汉生命之美科技有限公司