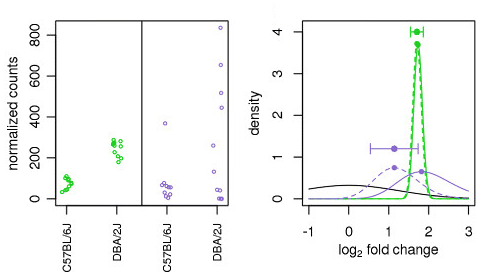
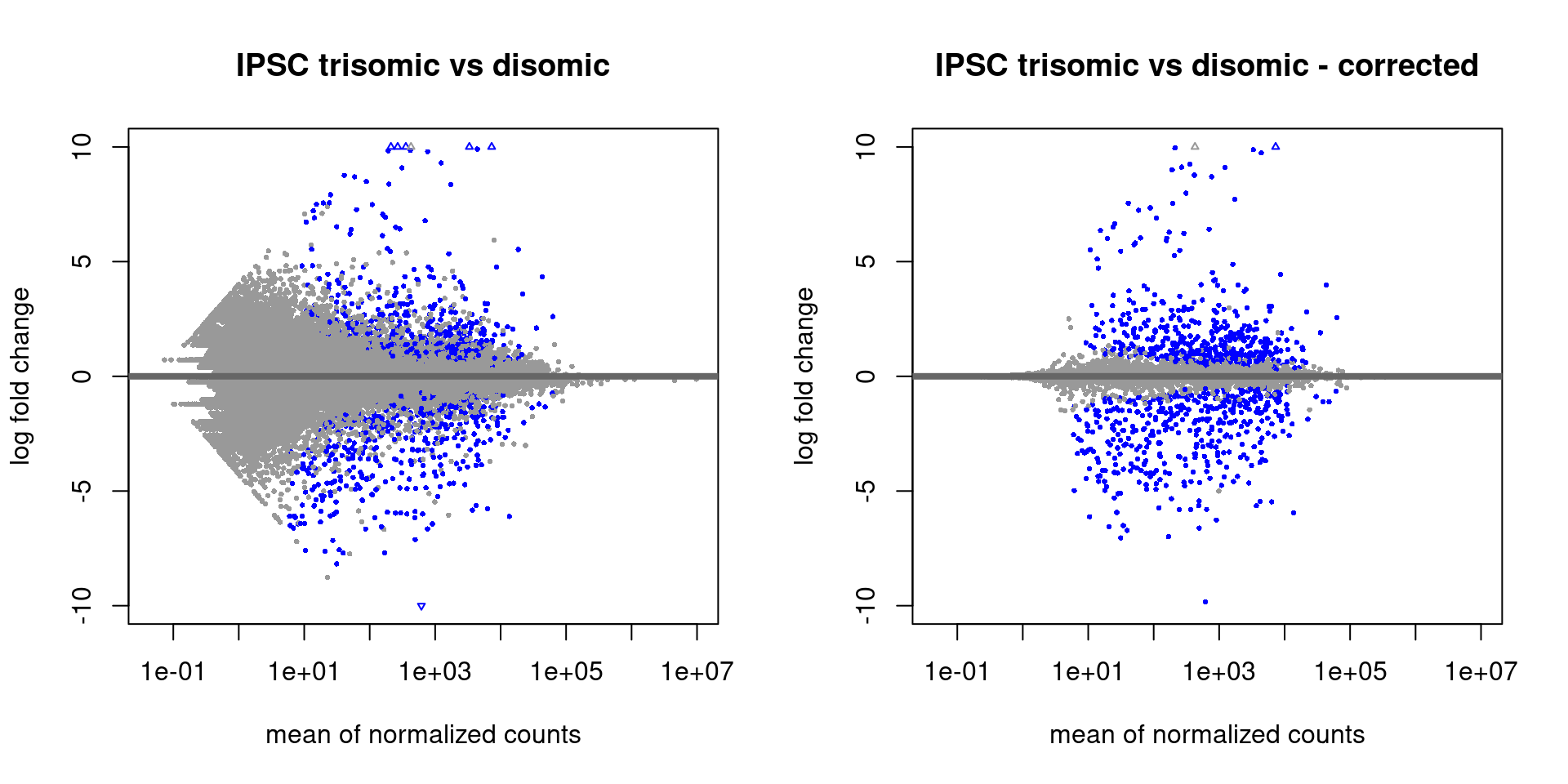
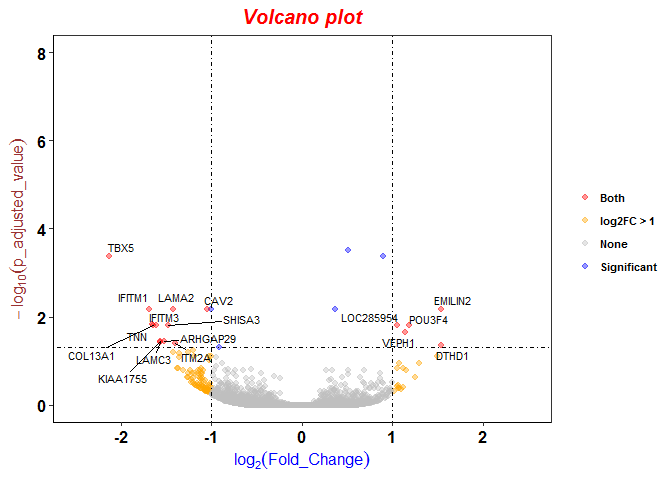
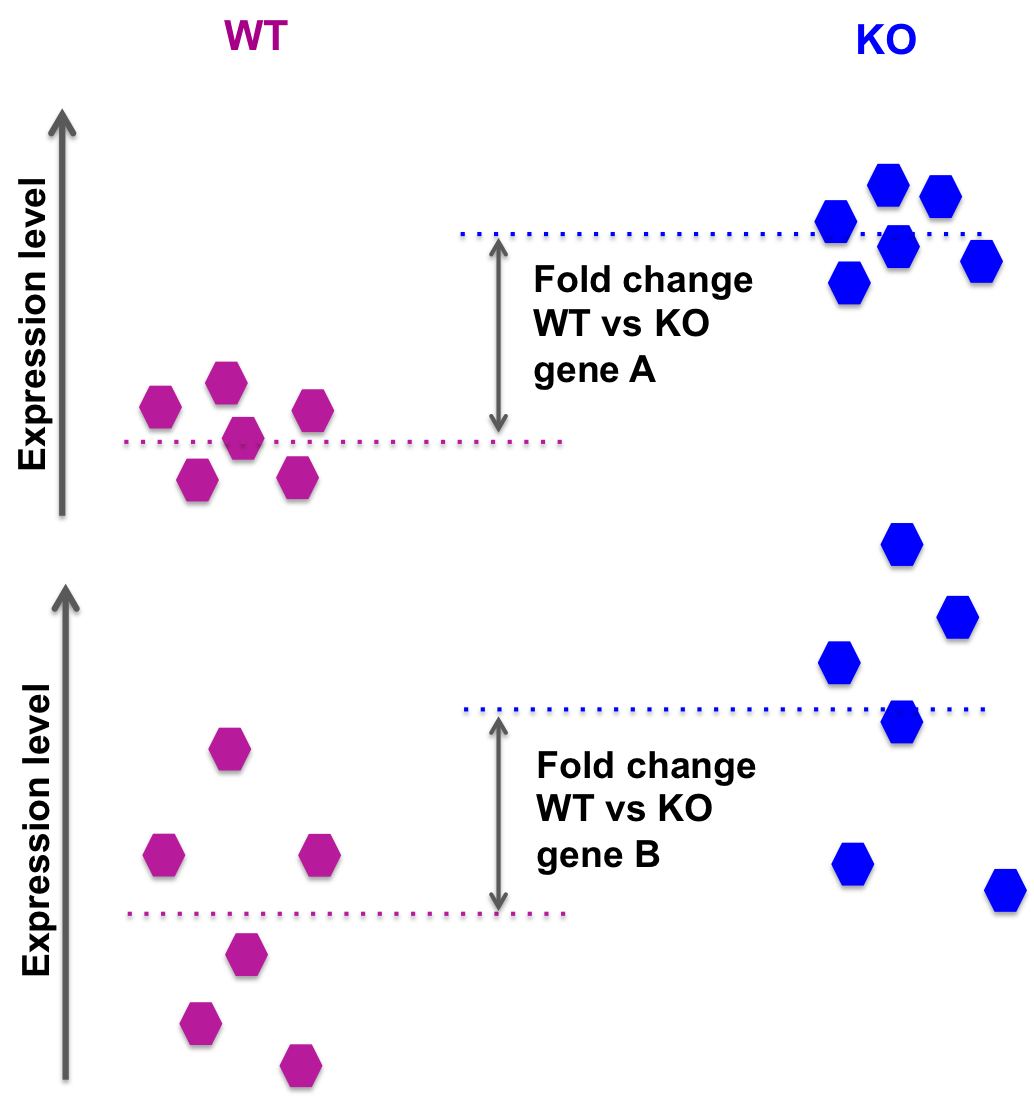
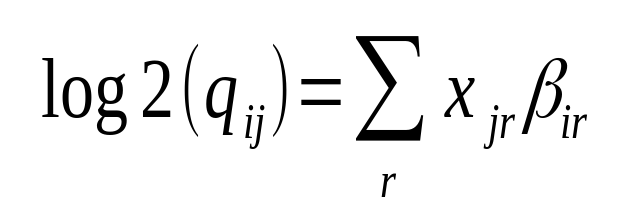Expression Profiling of Major Histocompatibility and Natural Killer Complex Genes Reveals Candidates for Controlling Risk of Graft versus Host Disease | PLOS ONE
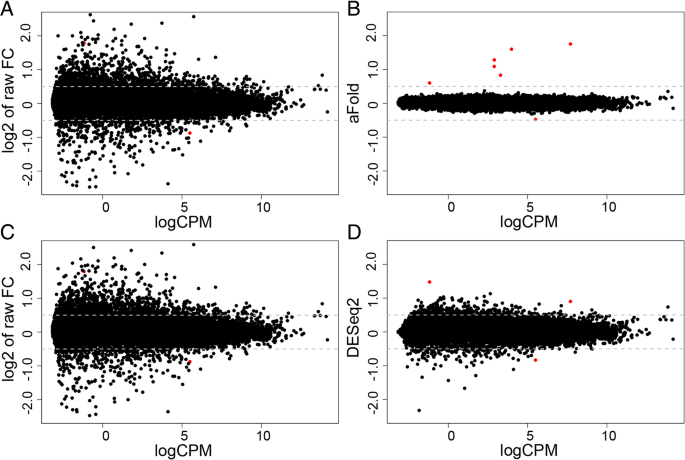
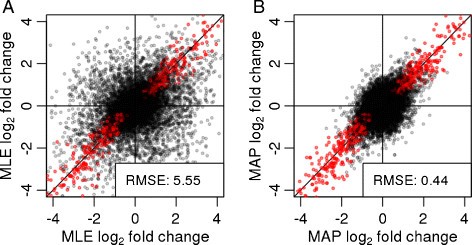
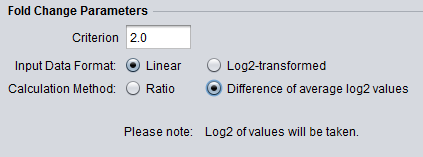
aFold – using polynomial uncertainty modelling for differential gene expression estimation from RNA sequencing data | BMC Genomics | Full Text
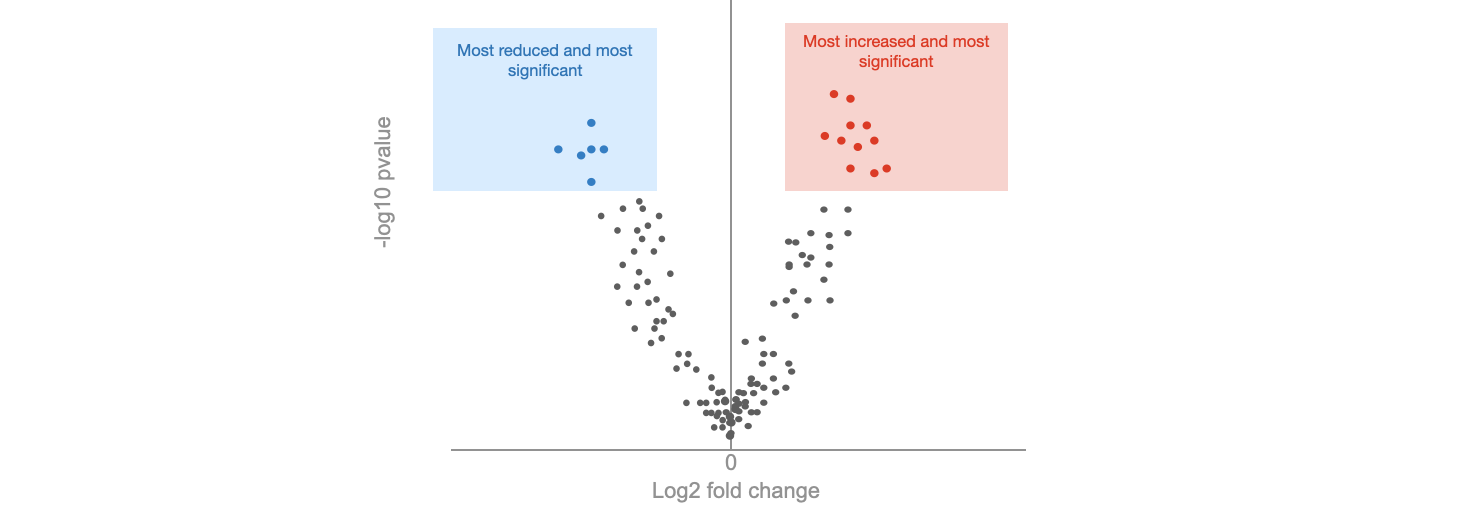
Integrated Analysis of Gene Expression, CpG Island Methylation, and Gene Copy Number in Breast Cancer Cells by Deep Sequencing | PLOS ONE
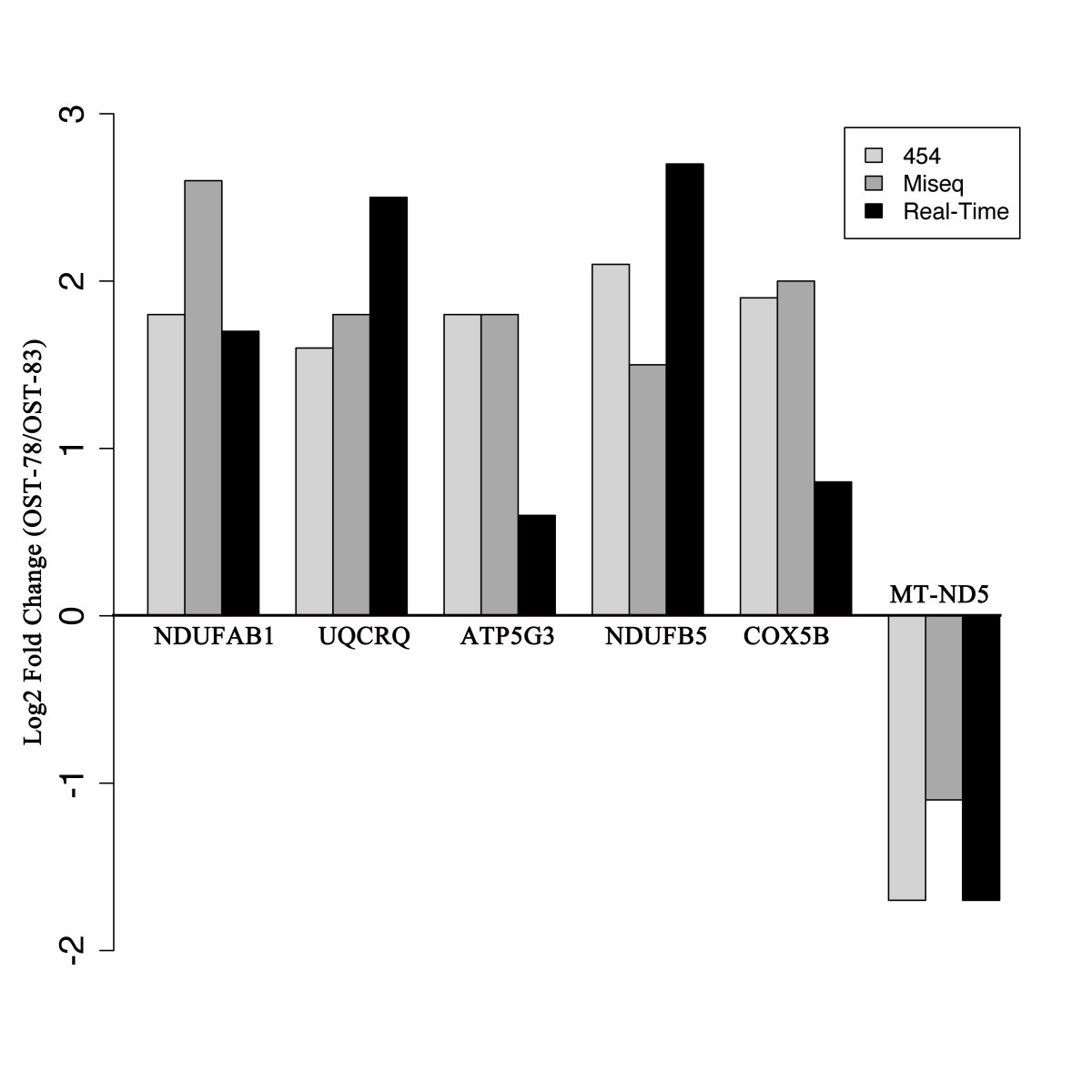
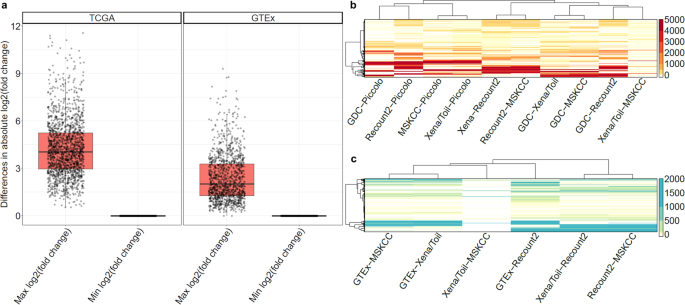
A platform independent RNA-Seq protocol for the detection of transcriptome complexity | BMC Genomics | Full Text

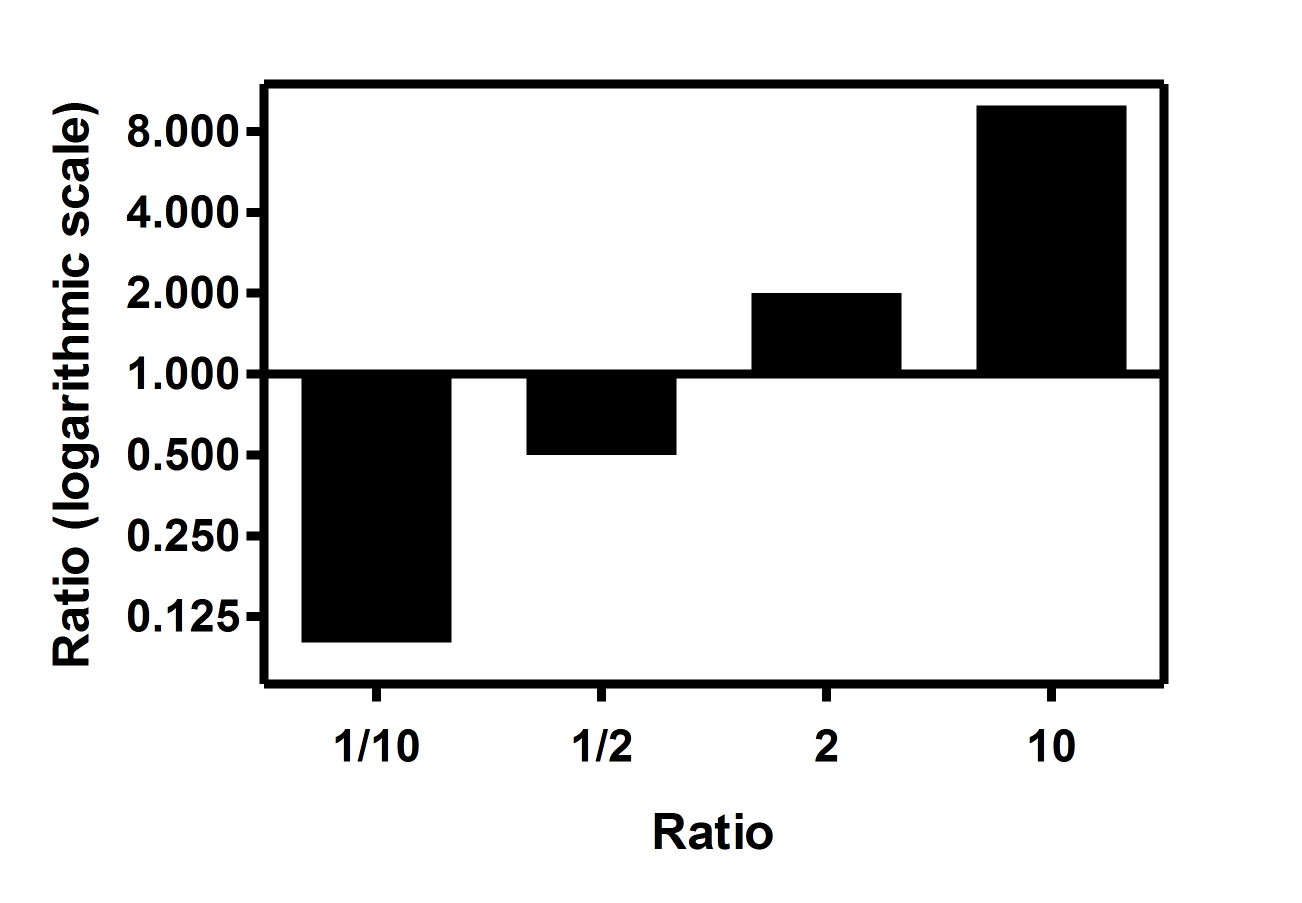
Log2-fold change values determined by qPCR for 11 markers identified as... | Download Scientific Diagram
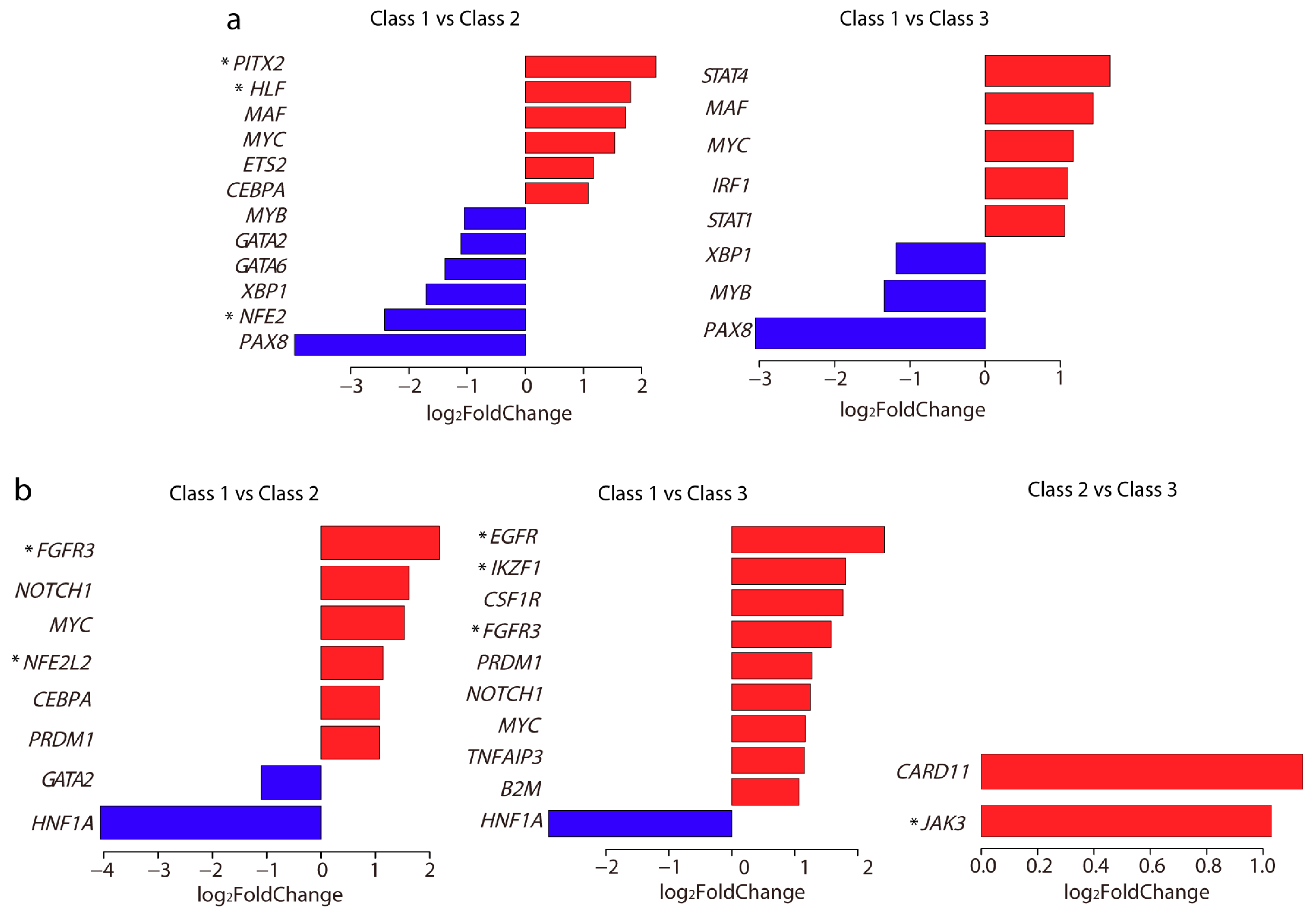
IJMS | Free Full-Text | Methylation-Based Classification of Cervical Squamous Cell Carcinoma into Two New Subclasses Differing in Immune-Related Gene Expression